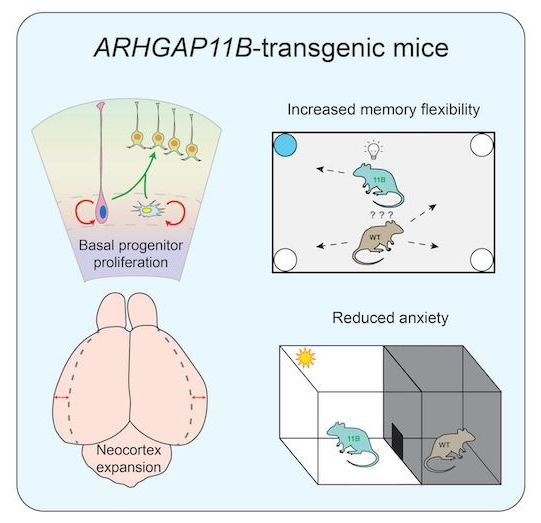
Generation of an ARHGAP11B-transgenic mouse line.
To generate a stable transgenic mouse line that expresses an ARHGAP11B protein, we first determined the temporal and spatial expression patterns of human ARHGAP11A and human ARHGAP11B mRNAs by qPCR of foetal human neocortical tissue at various developmental stages (gestational weeks 12–21; Fig EV1A and B) and by analysing previously published RNA-seq data sets of defined isolated NPC and neuron populations (Florio et al, 2015) (Fig EV1C and D), respectively. As the expression patterns of ARHGAP11A and ARHGAP11B mRNAs were found to be similar, we decided to generate the transgenic mouse line by converting one allele of the mouse Arhgap11a gene into a mutant mouse ARHGAP11B gene (mARHGAP11B), using the CRISPR/Cas9 genome editing technology (for details, see Materials and Methods). In m ARHGAP11B, the 55 nucleotides of Arhgap11a that in humans would be deleted from the ARHGAP11B mRNA by splicing using the new splice-donor site are replaced by the 141 nucleotides encoding the human-specific 47-amino acid sequence plus three nucleotides to generate a translational stop codon (Fig EV1E). Unless indicated otherwise, the ARHGAP11B-transgenic mice obtained (referred to as 11B mice hereafter) were used as heterozygous animals, that is, with one mouse Arhgap11a allele being replaced by m ARHGAP11B. The resulting ARHGAP11B protein will be expressed in developing mouse neocortex under the control of the native mouse Arhgap11a promotor.