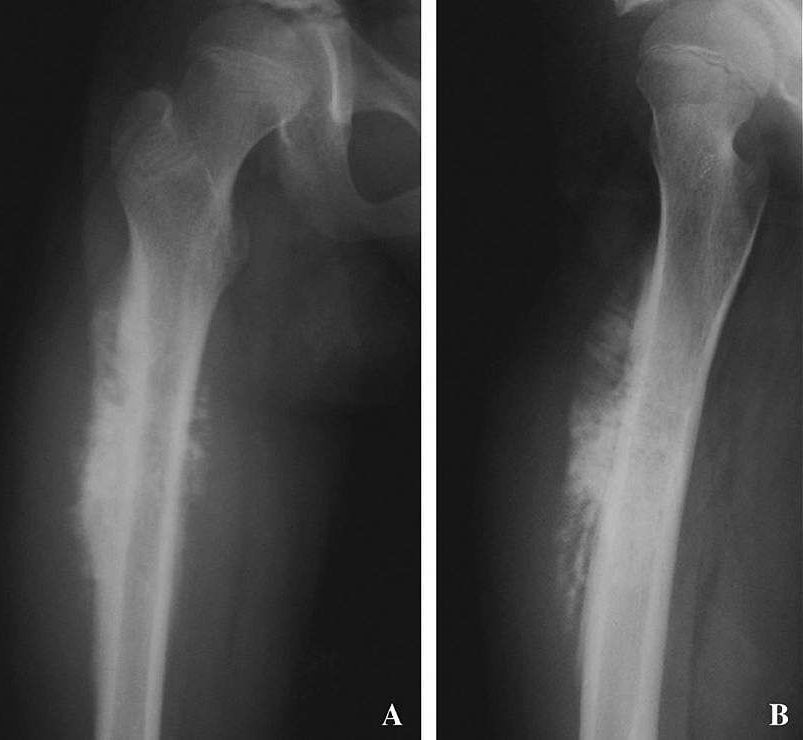
Andrew Sinclair does not have anything to lose. He takes a number of drugs including the anti-diabetic medication metformin, given to him by his son David, the renowned Australian biologist and professor of genetics at Harvard Medical School, to combat the ill-effects of ageing.
David Sinclair says his father remains in good health, travelling, socialising and exercising with the energy of a man far younger than his 80 years.
David Sinclair will discuss why ageing should be classified as a disease at the Festival of Dangerous Ideas.
Renowned Australian scientist David Sinclair says we have all the information to be young again, “if we can just flip the switch”.