The landmark decision has been eagerly awaited by millions but will be hotly contested by some experts doubt the drug’s effectiveness.


Summary: Study identifies genes that become activated in the brain prior to the initiation of severe repetitive behaviors associated with addiction, ASD, and schizophrenia.
Source: MIT
Extreme repetitive behaviors such as hand-flapping, body-rocking, skin-picking, and sniffing are common to a number of brain disorders including autism, schizophrenia, Huntington’s disease, and drug addiction. These behaviors, termed stereotypies, are also apparent in animal models of drug addiction and autism.

Check out this short educational video in which I explain some super exciting research in the area of nanotechnology: gigadalton-scale DNA origami! I specifically discuss a journal article by Wagenbauer et al. titled “Gigadalton-scale shape-programmable DNA assemblies”.
Here, I explain an exciting nanotechnology paper “Gigadalton-scale shape-programmable DNA assemblies” (https://doi.org/10.1038/nature24651).
Though I am not involved in this research myself, I have worked in adjacent areas such as synthetic biology, nanotechnology-based tools for neuroscience, and gene therapy. I am endlessly fascinated by DNA origami and would love to use it in my own research at some point in the future.
I am a PhD candidate at Washington University in St. Louis and the CTO of the startup company Conduit Computing. I am also a published science fiction writer and a futurist. To learn more about me, check out my website: https://logancollinsblog.com/.

Summary: Researchers have identified significant differences in gene activity between the anterior and posterior areas of the hippocampus. Genes associated with depression and other mood disorders are more active in the anterior hippocampus, while genes linked to cognitive disorders, such as ASD, are more active in the posterior hippocampus.
Source: UT Southwestern Medical Center.
A study of gene activity in the brain’s hippocampus, led by UT Southwestern researchers, has identified marked differences between the region’s anterior and posterior portions.

Enrolling Tens Of Thousands Of Dogs, In 10-Year Study, To Unlock Healthy Aging Secrets — Dr Matt Kaeberlein, Founder / Co-Director, The Dog Aging Project, Professor, University of Washington, joins me on Progress, Potential, And Possibilities Nathan Shock Centers #Rapamycin #Dogs #Aging #Longevity #Healthspan #Geroscience.
#MitochondrialDisease
Dr. Matt Kaeberlein is Professor of Pathology, Adjunct Professor of Genome Sciences, and Adjunct Professor of Oral Health Sciences, at the University of Washington.
Dr. Kaeberlein received his PhD from MIT in Biology, did his post-doc in the Department of Genome Sciences, University of Washington, and his research interests are focused on basic mechanisms of aging in order to facilitate translational interventions that promote healthspan and improve quality of life.
Dr. Kaeberlein has published nearly 200 papers in top scientific journals and has been recognized by several prestigious awards, including a Breakthroughs in Gerontology Award, an Alzheimer’s Association Young Investigator Award, an Ellison Medical Foundation New Scholar in Aging Award, a Murdock Trust Award, a Pioneer in Aging Award, and the Vincent Cristofalo Rising Star in Aging Research.
Dr. Kaeberlein’s contributions have also been recognized with Fellow status in the American Association for the Advancement of Science, the American Aging Association, and the Gerontological Society of America.
Dr. Kaeberlein is a past President of the American Aging Association and has served on their Executive Committee and Board of Directors since 2012. He has also served as a member of the Board of Directors for the Federation of American Societies for Experimental Biology and is currently the Chair of the Biological Sciences Section of the Gerontological Society of America.
Dr. Kaeberlein serves on the editorial boards for several journals, including Science and eLife. Dr. Kaeberlein’s scientific discoveries have generated substantial public interest, with featured stories in major media outlets including appearing on the front page of the New York Times, the Today Show, CNN, the UK Telegraph, Popular Science, Time Magazine, Scientific American, NPR, USA Today, National Geographic, and many others.
In addition to his primary appointments, Dr. Kaeberlein is the co-Director of the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging, the Founding Director of the Healthy Aging and Longevity Research Institute at the University of Washington, and Founder and Co-Director of the Dog Aging Project.

The Tsimane, an indigenous people who live in the Bolivian peripheries of the Amazon rainforest, lead lives that are very different to ours. They seem to be much healthier for it.
This tribal and largely isolated population of forager-horticulturalists still lives today by traditional ways of farming, hunting, gathering, and fishing – continuing the practices of their ancestors, established in a time long before industrialization and urbanization transformed most of the world.
For the Tsimane, the advantages are considerable. A study published in 2017 found that they effectively have the healthiest hearts in the world, with the lowest reported levels of coronary artery disease of any population ever recorded.
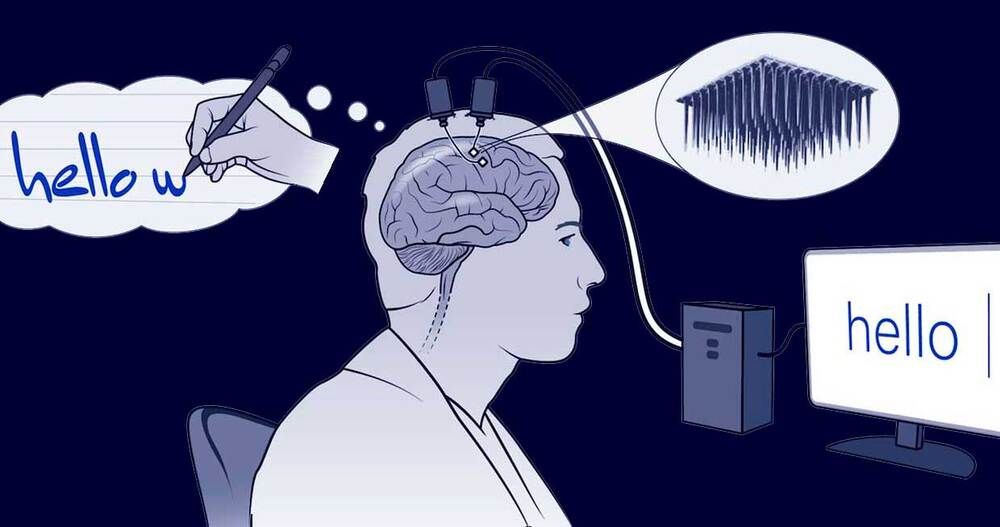
Erika Woodrum/HHMI/NatureTwo tiny arrays of implanted electrodes relayed information from the brain area that controls the hands and arms to an algorithm, which translated it into letters that appeared on a screen.

At Boston University, a team of researchers is working to better understand how language and speech is processed in the brain, and how to best rehabilitate people who have lost their ability to communicate due to brain damage caused by a stroke, trauma, or another type of brain injury. This type of language loss is called aphasia, a long-term neurological disorder caused by damage to the part of the brain responsible for language production and processing that impacts over a million people in the US.
“It’s a huge problem,” says Swathi Kiran, director of BU’s Aphasia Research Lab, and College of Health & Rehabilitation Sciences: Sargent College associate dean for research and James and Cecilia Tse Ying Professor in Neurorehabilitation. “It’s something our lab is working to tackle at multiple levels.”
For the last decade, Kiran and her team have studied the brain to see how it changes as people’s language skills improve with speech therapy. More recently, they’ve developed new methods to predict a person’s ability to improve even before they start therapy. In a new paper published in Scientific Reports, Kiran and collaborators at BU and the University of Texas at Austin report they can predict language recovery in Hispanic patients who speak both English and Spanish fluently—a group of aphasia patients particularly at risk of long-term language loss—using sophisticated computer models of the brain. They say the breakthrough could be a game changer for the field of speech therapy and for stroke survivors impacted by aphasia.
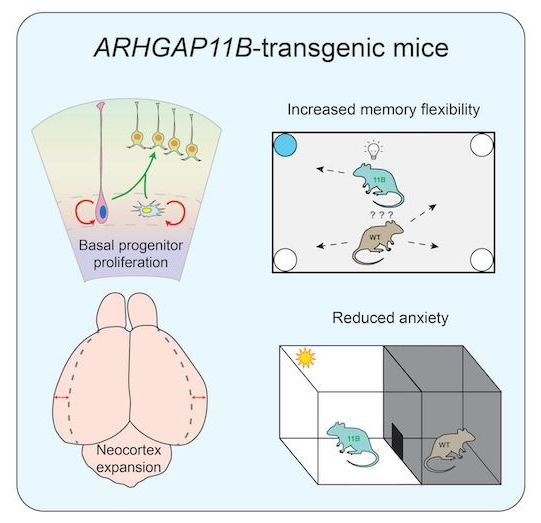
Generation of an ARHGAP11B-transgenic mouse line.
To generate a stable transgenic mouse line that expresses an ARHGAP11B protein, we first determined the temporal and spatial expression patterns of human ARHGAP11A and human ARHGAP11B mRNAs by qPCR of foetal human neocortical tissue at various developmental stages (gestational weeks 12–21; Fig EV1A and B) and by analysing previously published RNA-seq data sets of defined isolated NPC and neuron populations (Florio et al, 2015) (Fig EV1C and D), respectively. As the expression patterns of ARHGAP11A and ARHGAP11B mRNAs were found to be similar, we decided to generate the transgenic mouse line by converting one allele of the mouse Arhgap11a gene into a mutant mouse ARHGAP11B gene (mARHGAP11B), using the CRISPR/Cas9 genome editing technology (for details, see Materials and Methods). In m ARHGAP11B, the 55 nucleotides of Arhgap11a that in humans would be deleted from the ARHGAP11B mRNA by splicing using the new splice-donor site are replaced by the 141 nucleotides encoding the human-specific 47-amino acid sequence plus three nucleotides to generate a translational stop codon (Fig EV1E). Unless indicated otherwise, the ARHGAP11B-transgenic mice obtained (referred to as 11B mice hereafter) were used as heterozygous animals, that is, with one mouse Arhgap11a allele being replaced by m ARHGAP11B. The resulting ARHGAP11B protein will be expressed in developing mouse neocortex under the control of the native mouse Arhgap11a promotor.