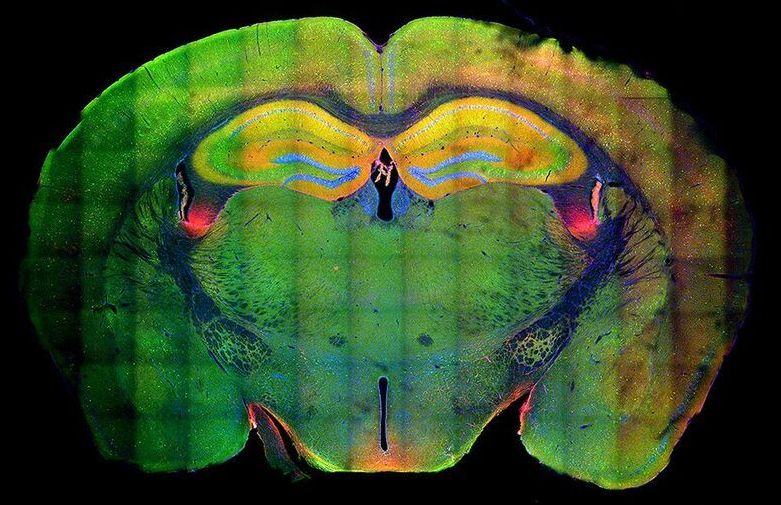
Researchers see structural changes in genetic material that allow memories to strengthen when remembered.


In a review published in the journal *Science*, Jain and Steele Laboratories colleagues Hadi T. Nia, PhD, and Lance L. Munn, PhD, describe four distinct physical hallmarks of cancer that affect both cancer cells and the tumor microenvironment, contributing to both tumor growth and the development of resistance to powerful cancer drugs.
One widely accepted model of cancer holds that a normal cell goes rogue because of genetic mutations or an environmental insult. In this model, the altered cell starts replicating out of control and takes over normal tissues, displaying eight hallmarks that include the ability to promote and sustain the growth of tumors, evade immune system attempts to suppress growth, stimulate blood flow to tumors and both invade local tissues and metastasize (spread) elsewhere in the body.
But this model fails to take into account how physical processes affect tumor progression and treatment, say the authors. In addition to the aforementioned eight biological hallmarks of cancer proposed by Robert Weinberg, PhD, from MIT, and Douglas Hanahan, PhD, from the Swiss Federal Institute of Technology in Lausanne, Jain and colleagues propose adding four distinct physical hallmarks that capture the biomechanical abnormalities in tumors: elevated solid stress; elevated interstitial fluid pressure; increased stiffness and altered material properties; and altered tissue micro-architecture.
Three decades of research in the Steele Laboratories led to the discovery and clinical translation of the first two hallmarks. “Solid stresses are created as proliferating and migrating cells push and stretch solid components of the surrounding tissue. They are large enough to compress blood and lymphatic vessels in and around tumors, impairing blood flow and the delivery of oxygen, drugs and immune cells,” Jain says.
Elevated interstitial fluid pressure is caused by abnormally permeable blood vessels in tumors leaking blood plasma into tissues surrounding the tumor, and by insufficient drainage of lymphatic fluid. The interstitial fluid carries various growth factors with it, causing edema (swelling), elution (release) of drugs and growth factors, and facilitating cancer invasion of local and distant tissues.
Increased stiffness is caused by the deposition of cellular matrix (scaffolding) and remodeling of tissues. This stiffness has traditionally been used as a diagnostic marker for tumor growth, and more recently it has come to be recognized as a marker for prognosis. Increased stiffness activates signaling pathways that promote proliferation, invasiveness and metastasis of cancer cells, Jain explains.
“Finally, when normal tissue architecture is disrupted by cancer growth and invasion, micro-architecture is altered,” he says. “Stromal (supporting) cells, cancer cells and extracellular matrix adopt new organization. This changes the interactions between an individual cell and its surrounding matrix and cells, which affects signaling pathways associated with invasion and metastasis.”
Jain says that with the review article in Science, he and his colleagues hope to bridge the gap between the physical and biological sciences “by exploring the biological origins and repercussions of the physical hallmarks of cancer from the perspectives of cancer biologists and oncologists — who work to understand and overcome the physical abnormalities at the bench and in the clinic — and from the perspectives of physicists and engineers who develop new models and strategies for research, diagnosis and treatment.”
An evolving understanding of cancer that incorporates the physical properties of tumors and their surrounding tissues into existing biologic and genetic models can direct cancer researchers down previously uncharted avenues, potentially leading to new drugs and new treatment strategies, say investigators from Massachusetts General Hospital (MGH), Harvard Medical School (HMS) and the Ludwig Center at HMS.
“We believe that progress in cancer research relies on close collaboration between cancer biologists, oncologists, physical scientists and engineers. A comprehensive understanding of the physical hallmarks of cancer requires a rigorous and broad perspective spanning the physical and biological sciences,” says Rakesh K. Jain, PhD, an investigator in the Edwin L. Steele Laboratories in the Department of Radiation Oncology at MGH and HMS.
In a review published in the journal Science, Jain and Steele Laboratories colleagues Hadi T. Nia, PhD, and Lance L. Munn, PhD, describe four distinct physical hallmarks of cancer that affect both cancer cells and the tumor microenvironment, contributing to both tumor growth and the development of resistance to powerful cancer drugs.

Li’l Kurt is a genetic marvel.
In an effort to increase genetic diversity among horses, scientists have gone sci-fi and used frozen 40-year-old cells to create Kurt, the very first clone of a Przewalski’s horse.
🐴 You love badass animals. So do we. Let’s nerd out over them together.

Mice who ate a diet high in fat and cholesterol were more likely to see their hair turn from black to white and experience hair loss. The diet also appeared to cause inflammation of the skin.
In the first stage of the study, the researchers genetically modified mice to develop atherosclerosis, a condition in which fat deposits form in the arteries.
They then fed mice either a Western diet high in fat and cholesterol or untreated rat chow from the age of 12 to 20 weeks. As expected, the mice who consumed the Western diet saw their hair turn white and fall out, and develop skin lesions. And the longer the mice ate the diet, the worse their symptoms became. By week 36, three quarters of the animals had skin lesions.

The great powerful guppy can essentially evolve 10 million times faster than usual. Which could lead to humans evolving faster too leading to a biological singularity.
Although natural selection is often viewed as a slow pruning process, a dramatic new field study suggests it can sometimes shape a population as fast as a chain saw can rip through a sapling. Scientists have found that guppies moved to a predator-free environment adapted to it in a mere 4 years—a rate of change some 10,000 to 10 million times faster than the average rates gleaned from the fossil record. Some experts argue that the 11-year study, described in today’s issue of Science,* may even shed light on evolutionary patterns that occur over eons.
A team led by evolutionary biologist David Reznick of the University of California, Riverside, scooped guppies from a waterfall pool brimming with predators in Trinidad’s Aripo River, then released them in a tributary where only one enemy species lurked. In as little as 4 years, male guppies in the predator-free tributary were already detectably larger and older at maturity when compared with the control population; 7 years later females were too. Guppies in the safer waters also lived longer and had fewer and bigger offspring.
The team next determined the rate of evolution for these genetic changes, using a unit called the darwin, or the proportional amount of change over time. The guppies evolved at a rate between 3700 and 45,000 darwins. For comparison, artificial-selection experiments on mice show rates of up to 200,000 darwins—while most rates measured in the fossil record are only 0.1 to 1.0 darwin. “It’s further proof that evolution can be very, very fast and dynamic,” says Philip Gingerich, a paleontologist at the University of Michigan, Ann Arbor. “It can happen on a time scale that’s as short as one generation—from us to our kids.”
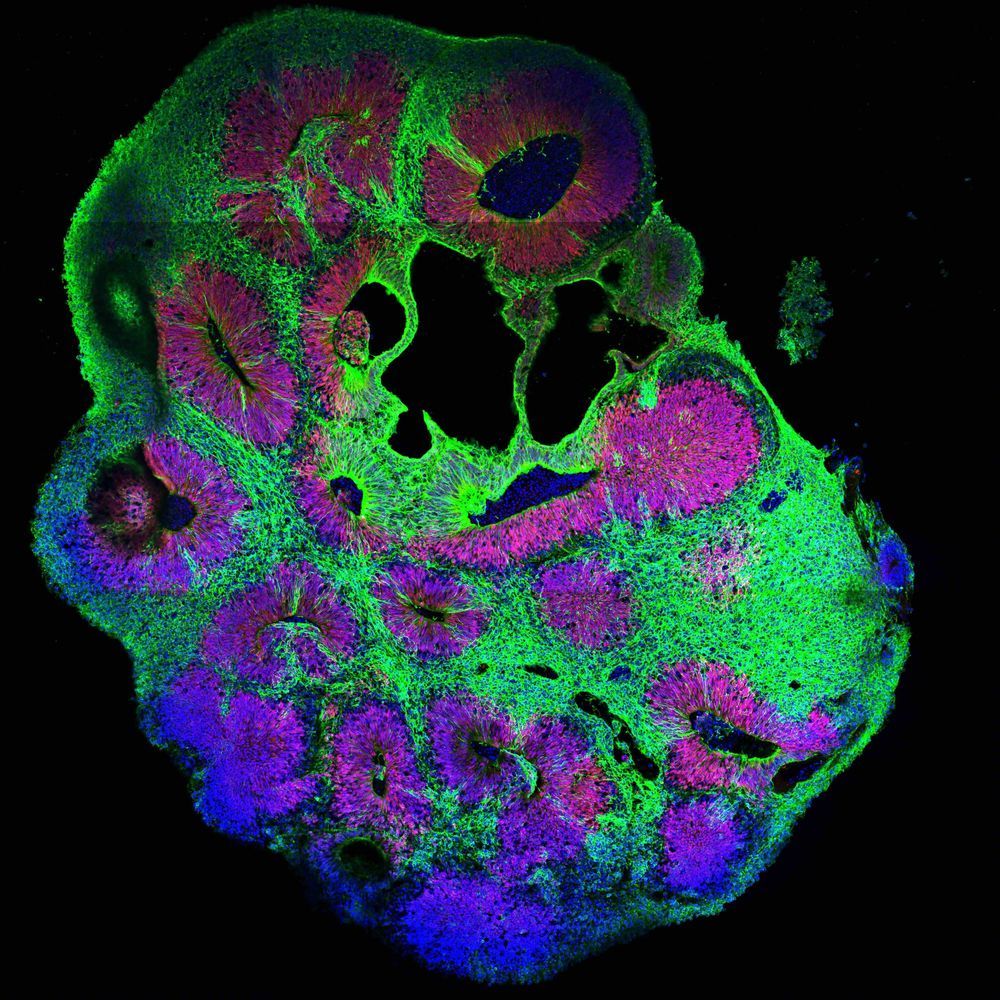
Many of the fundamental principles in biology and essentially all pathways regulating development were identified in so-called genetics screens. Originally pioneered in the fruit fly Drosophila and the nematode C. elegans, genetic screens involve inactivation of many genes one by one. By analyzing the consequences of gene loss, scientists can draw conclusions about its function. This way, for example, all genes required for formation of a brain can be identified.
Genetic screens can routinely be carried out in flies and worms. In humans, a wealth of knowledge exists about genetic disorders and the consequences of disease-relevant mutations, but their systematic analysis was impossible. Now, the Knoblich lab at IMBA has developed a groundbreaking technique allowing hundreds of genes to be analyzed in parallel in human tissue. They named the new technology CRISPR-LICHT and published their findings in the journal Science.
By using cerebral organoids, a 3D cell culture model for the human brain developed in Jürgen Knoblich’s group at IMBA, hundreds of mutations can now be analyzed for their role in the human brain using CRISPR-LICHT.
While our circadian body clock dictates our preferred rhythm of sleep or wakefulness, a relatively new concept—the epigenetic clock—could inform us about how swiftly we age, and how prone we are to diseases of old age.
People age at different rates, with some individuals developing both characteristics and diseases related to aging earlier in life than others. Understanding more about this so-called ‘biological age’ could help us learn more about how we can prevent diseases associated with age, such as dementia. Epigenetic markers control the extent to which genes are switched on and off across the different cell-types and tissues that make up a human body. Unlike our genetic code, these epigenetic marks change over time, and these changes can be used to accurately predict biological age from a DNA sample.
Now, scientists at the University of Exeter have developed a new epigenetic clock specifically for the human brain. As a result of using human brain tissue samples, the new clock is far more accurate than previous versions, that were based on blood samples or other tissues. The researchers hope that their new clock, published in Brain and funded by Alzheimer’s Society, will provide insight into how accelerated aging in the brain might be associated with brain diseases such as Alzheimer’s disease and other forms of dementia.

The coronavirus disease 2019 (COVID-19) pandemic does not affect everyone equally. While anyone can contract COVID-19, accumulating data suggest that older people or those with pre-existing comorbidities are far more likely to have severe complications or die from the disease. While researchers scramble to unravel the mechanisms of action underlying the disease’s wide-ranging effects, news that the disease hits older people hardest has been received without demur: it is widely accepted that to be old is to be fragile. Indeed, even in so-called normal times, everyone expects more things break as people age: bones, hearts, brains. In the context of the pandemic, being old is seen as just one more comorbidity.
It should not be.
We accept growing old and losing our vitality as an inevitability of life. To do so is to overlook the fact that ageing is, fundamentally, a plastic trait—influenced both by our genetic predispositions and many (controllable) environmental factors. Anecdotally we know this to be true: for some, being in their eighties means being confined to a wheelchair whereas for others, like Eileen Noble, who at 84 years old was the oldest runner in 2019’s London Marathon, it decidedly does not. The burgeoning field of biogerontology is now beginning to amass data in support of such observations. Single genetic mutations in evolutionarily conserved pathways across model organisms—ranging from fruit flies to mice—increase lifespan by up to 80%. Crucially, not only do these animals live longer, they also have a longer youthspan—the proportion of their lives in which they retain the trappings of youth such as peak mobility, immunity, and stress resilience.
Summary: A mutation in a gene associated with circadian rhythm extends the clock period, causing people to stay up late at night and sleep late in the mornings.
Source: UC Santa Cruz
A new study by researchers at UC Santa Cruz shows how a genetic mutation throws off the timing of the biological clock, causing a common sleep syndrome called delayed sleep phase disorder.

KENNEDY SPACE CENTER (FL), October 19, 2020 – The Center for the Advancement of Science in Space (CASIS) and the National Science Foundation (NSF) announced three flight projects that were selected as part of a joint solicitation focused on leveraging the International Space Station (ISS) U.S. National Laboratory to further knowledge in the fields of tissue engineering and mechanobiology. Through this collaboration, CASIS, manager of the ISS National Lab, will facilitate hardware implementation, in-orbit access, and astronaut crew time on the orbiting laboratory. NSF invested $1.2 million in the selected projects, which are seeking to advance fundamental science and engineering knowledge for the benefit of life on Earth.
This is the third collaborative research opportunity between CASIS and NSF focused on tissue engineering. Fundamental science is a major line of business for the ISS National Lab, and by conducting research in the persistent microgravity environment offered by the orbiting laboratory, NSF and the ISS National Lab will drive new advances that will bring value to our nation and spur future inquiries in low Earth orbit.
Microgravity affects organisms—from viruses and bacteria to humans, inducing changes such as altered gene expression and DNA regulation, changes in cellular function and physiology, and 3D aggregation of cells. Spaceflight is advancing research in the fields of pharmaceutical research, disease modeling, regenerative medicine, and many other areas within the life sciences. The selected projects will utilize the ISS National Lab and its unique environment to advance fundamental and transformative research that integrates engineering and life sciences.