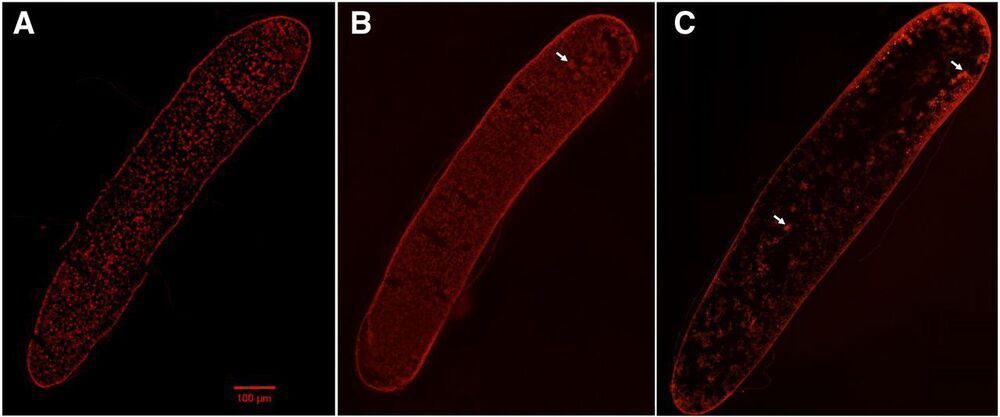
The honeybee (Apis mellifera) is an important insect pollinator of wild flowers and crops, playing critical roles in the global ecosystem. Additionally, the honeybee serves as an ideal social insect model. Therefore, functional studies on honeybee genes are of great interest. However, until now, effective gene manipulation methods have not been available in honeybees. Here, we reported an improved CRISPR/Cas9 gene-editing method by microinjecting sgRNA and Cas9 protein into the region of zygote formation within 2 hr after queen oviposition, which allows one-step generation of biallelic knockout mutants in honeybee with high efficiency. We first targeted the Mrjp1 gene. Two batches of honeybee embryos were collected and injected with Mrjp1 sgRNA and Cas9 protein at the ventral cephalic side and the dorsal posterior side of the embryos, respectively. The gene-editing rate at the ventral cephalic side was 93.3%, which was much higher than that (11.8%) of the dorsal-posterior-side injection. To validate the high efficiency of our honeybee gene-editing system, we targeted another gene, Pax6, and injected Pax6 sgRNA and Cas9 protein at the ventral cephalic side in the third batch. A 100% editing rate was obtained. Sanger sequencing of the TA clones showed that 73.3% (for Mrjp1) and 76.9% (for Pax6) of the edited current-generation embryos were biallelic knockout mutants. These results suggest that the CRISPR/Cas9 method we established permits one-step biallelic knockout of target genes in honeybee embryos, thereby demonstrating an efficient application to functional studies of honeybee genes. It also provides a useful reference to gene editing in other insects with elongated eggs.