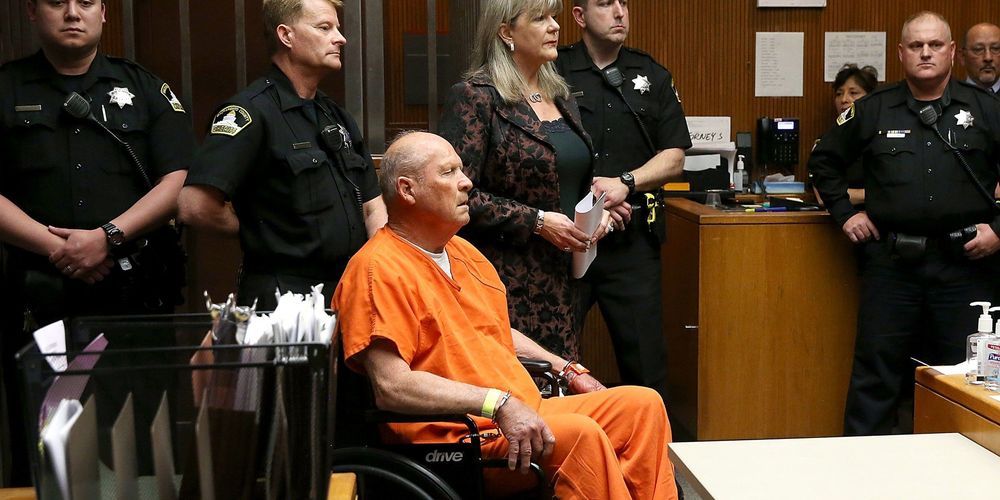Three hundred and sixty-six days ago, CeCe Moore woke up to the headline that would change her world: “Suspected Golden State Killer, East Area Rapist Arrested After Eluding Authorities for Decades.” Later that day, those authorities would hold a press conference in front of the Sacramento County District Attorney’s office to explain how, a day earlier, they had finally put handcuffs on the man believed to have committed a series of sadistic rapes and murders that spread terror through California for more than 40 years. But Moore didn’t have to tune in to know how they had done it. “I knew immediately they had cracked it with genetic genealogy and GEDmatch,” she says.
She knew it because at the time, Moore was working as the genetic genealogy researcher on the PBS show Finding Your Roots and had a consulting business helping adoptees find their biological parents. To aid her searches, she regularly logged on to GEDmatch, a public database where hobbyists upload results from consumer genetic testing companies like 23andMe and Ancestry to find relatives with shared DNA and to reverse-engineer their family trees. It had come to her attention that another genealogist on the site, Barbara Rae-Venter, had been uploading files that seemed out of place, and Moore suspected they came not from family members, but from crime scenes. But she had never imagined that one of them belonged to the man believed to be one of the most notorious serial killers in US history. “This was going to be huge,” she remembers telling people that day.
But not even Moore could have predicted just how huge it would become. In the year since the dramatic arrest of Joseph James DeAngelo, the alleged Golden State Killer, investigative genetic genealogy has emerged as the most powerful new crime-fighting tool since DNA itself. The technique has been used to identify suspects in more than 50 additional cases. Its vast potential to crack tens of thousands more has given rise to a lucrative new forensic science business, the formation of dedicated family-tree-building police units, and the first-ever home DNA kit marketing campaign to get people to send in their spit to solve crimes.