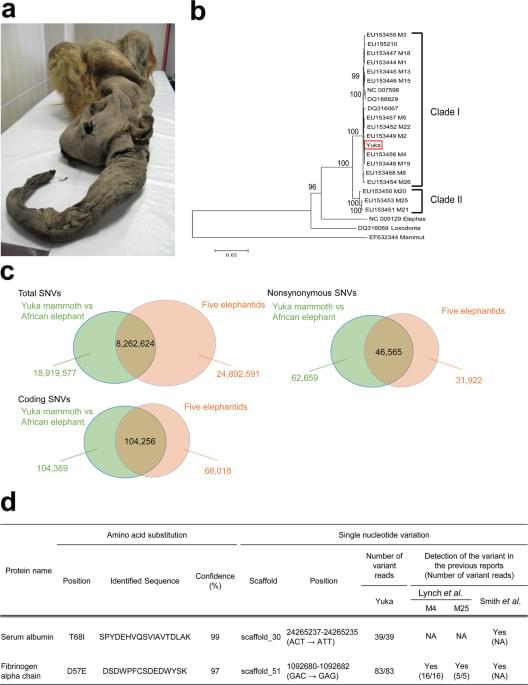
ORF Genetics in Iceland is growing 100,000 genetically engineered barley plants in a greenhouse measuring over 22 square feet (2 sq m) to create lab-grown meat.
This cutting-edge approach has the potential to lower prices, eliminate reliance on live animals in the lab-grown meat sector, and speed up the scaling-up process, according to BBC. And, with the fact that meat accounts for nearly 60 percent of all greenhouse gases from food production in mind, such a development could have far-reaching implications in the fight against climate change.