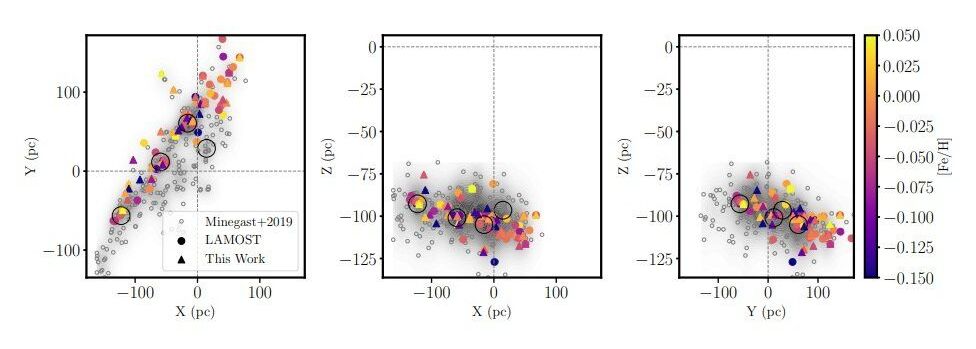
Stellar streams are long, thin filaments of stars orbiting galaxies, produced by the stretching action of tidal forces. For astronomers, observation of these structures could be crucial to test various galaxy formation models.
Located most likely some 420 light-years away in the Milky Way’s disk, Pisces–Eridanus (or Psc–Eri for short) is a cylindrically shaped stream of almost 1,400 identified stars distributed across about 2,300 light-years. Due to its relative proximity and population size, it is perceived as an excellent laboratory to study star formation and test theories of chemical and dynamical evolution of stellar systems.