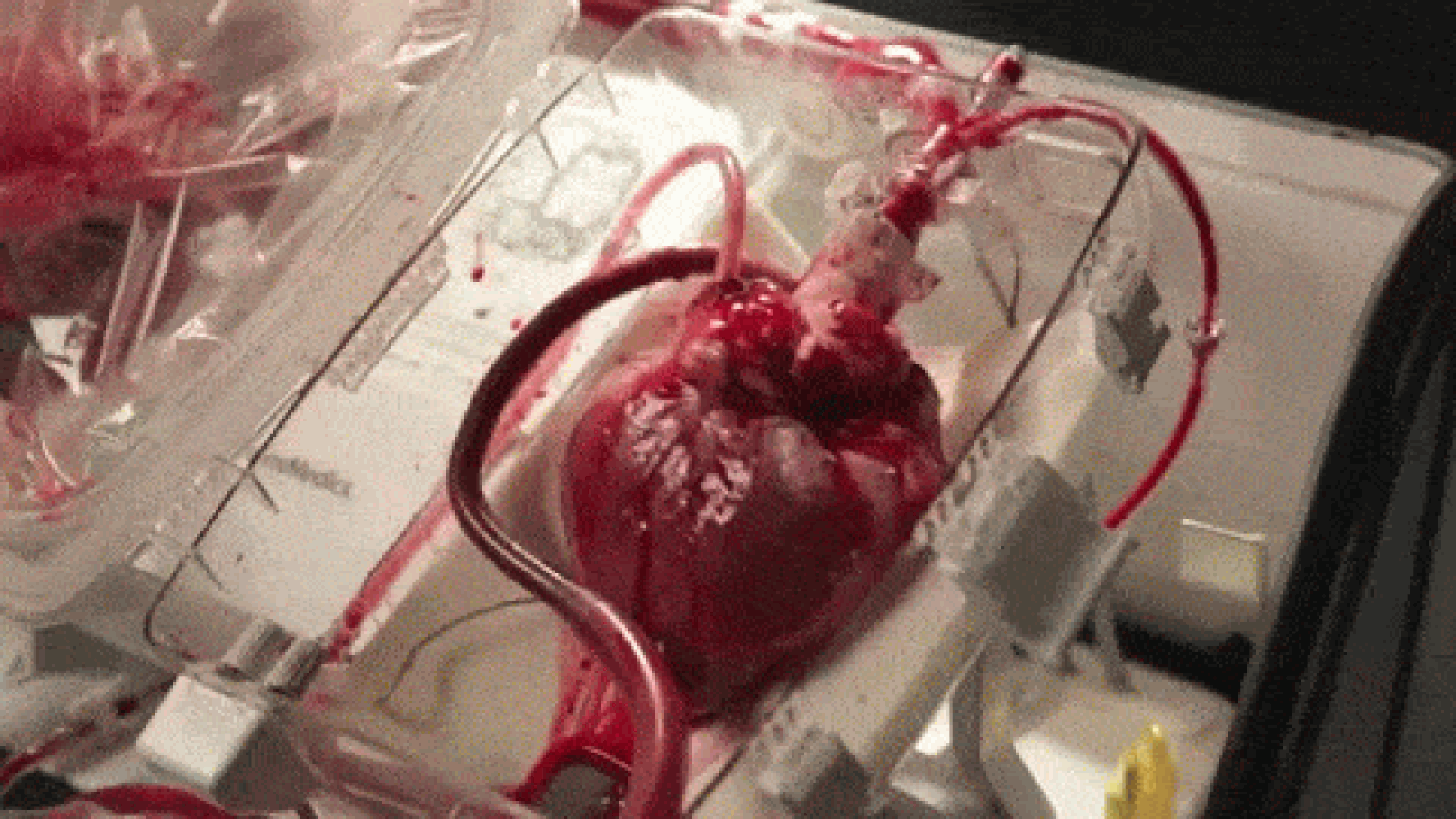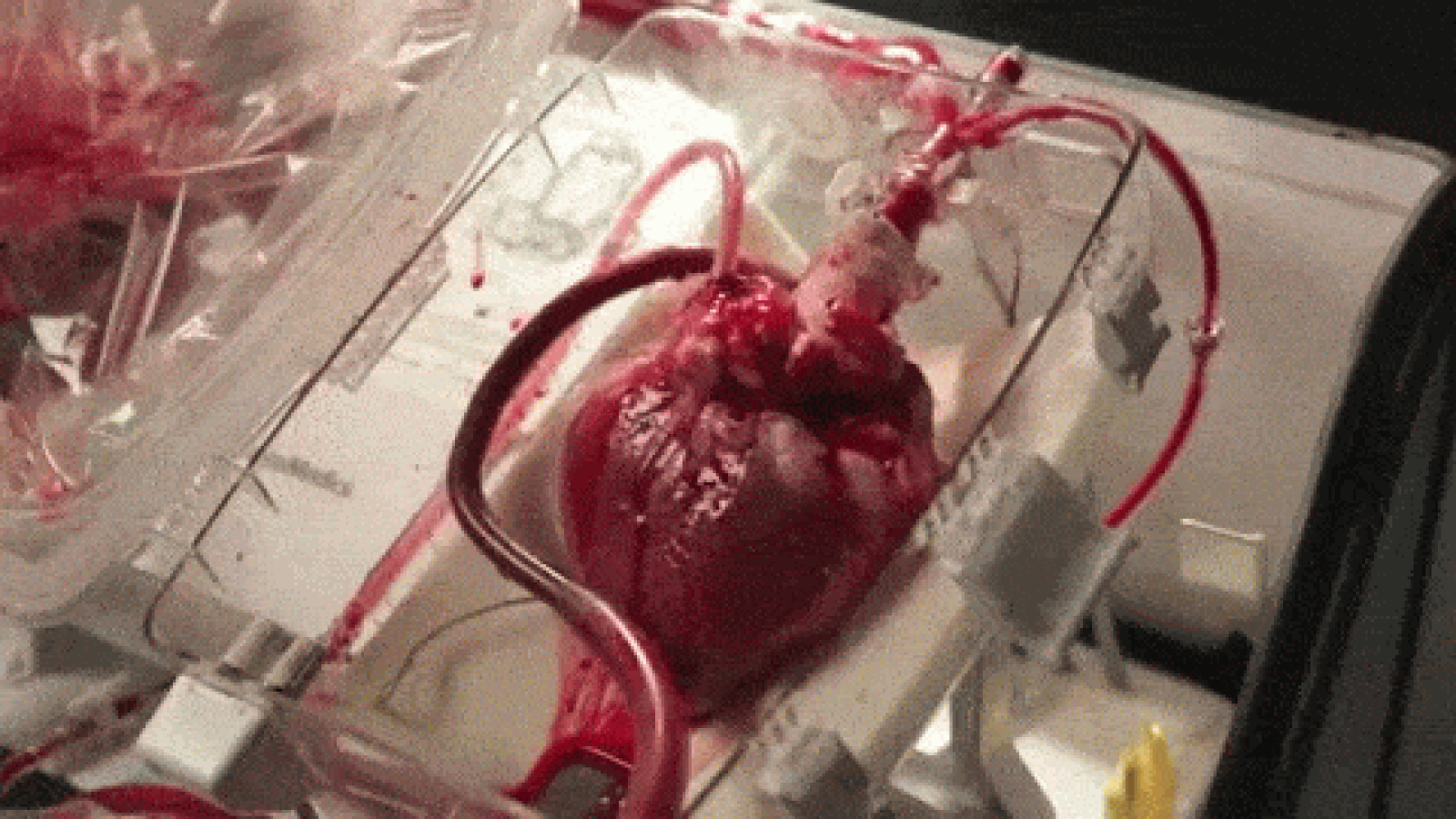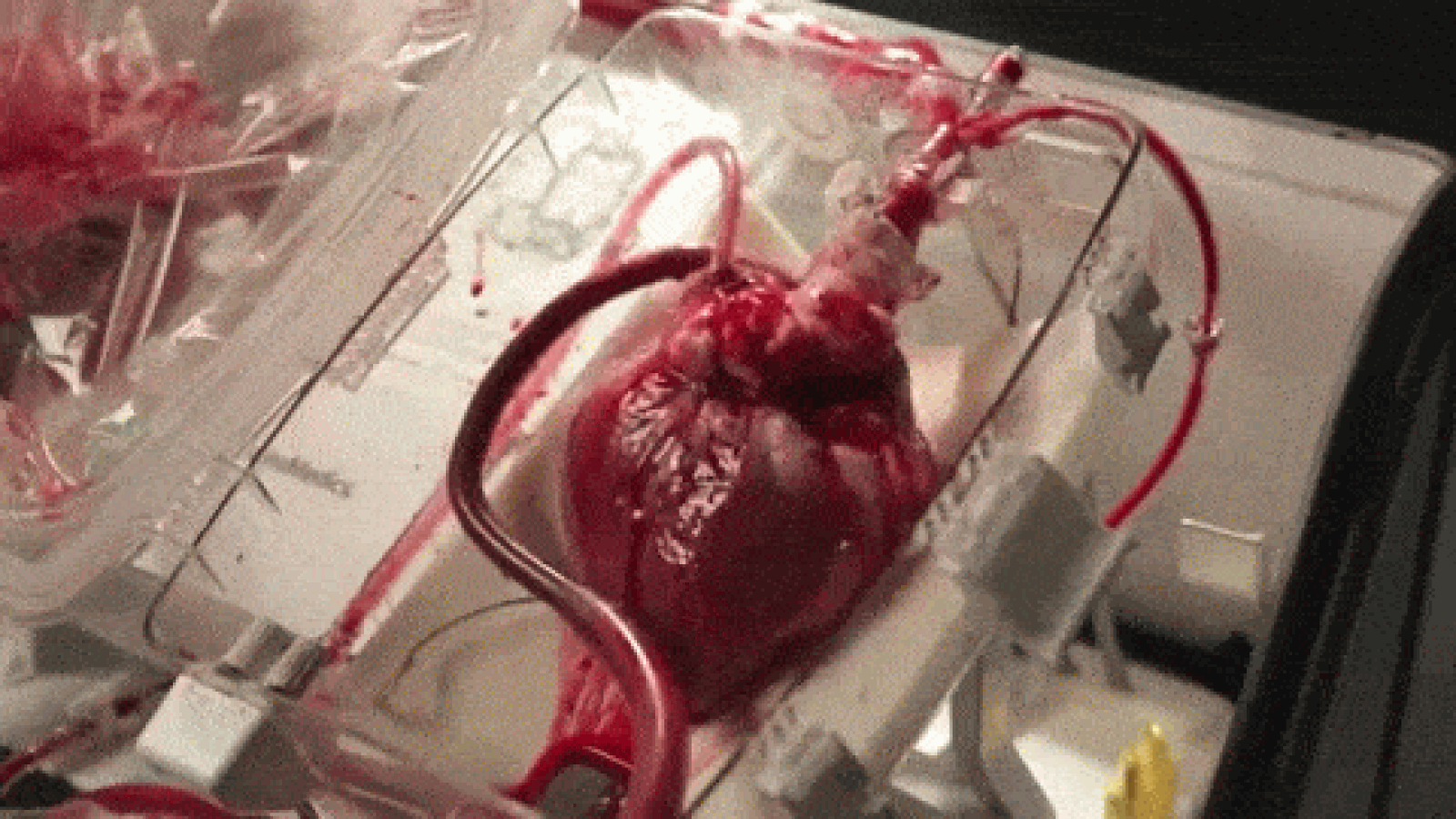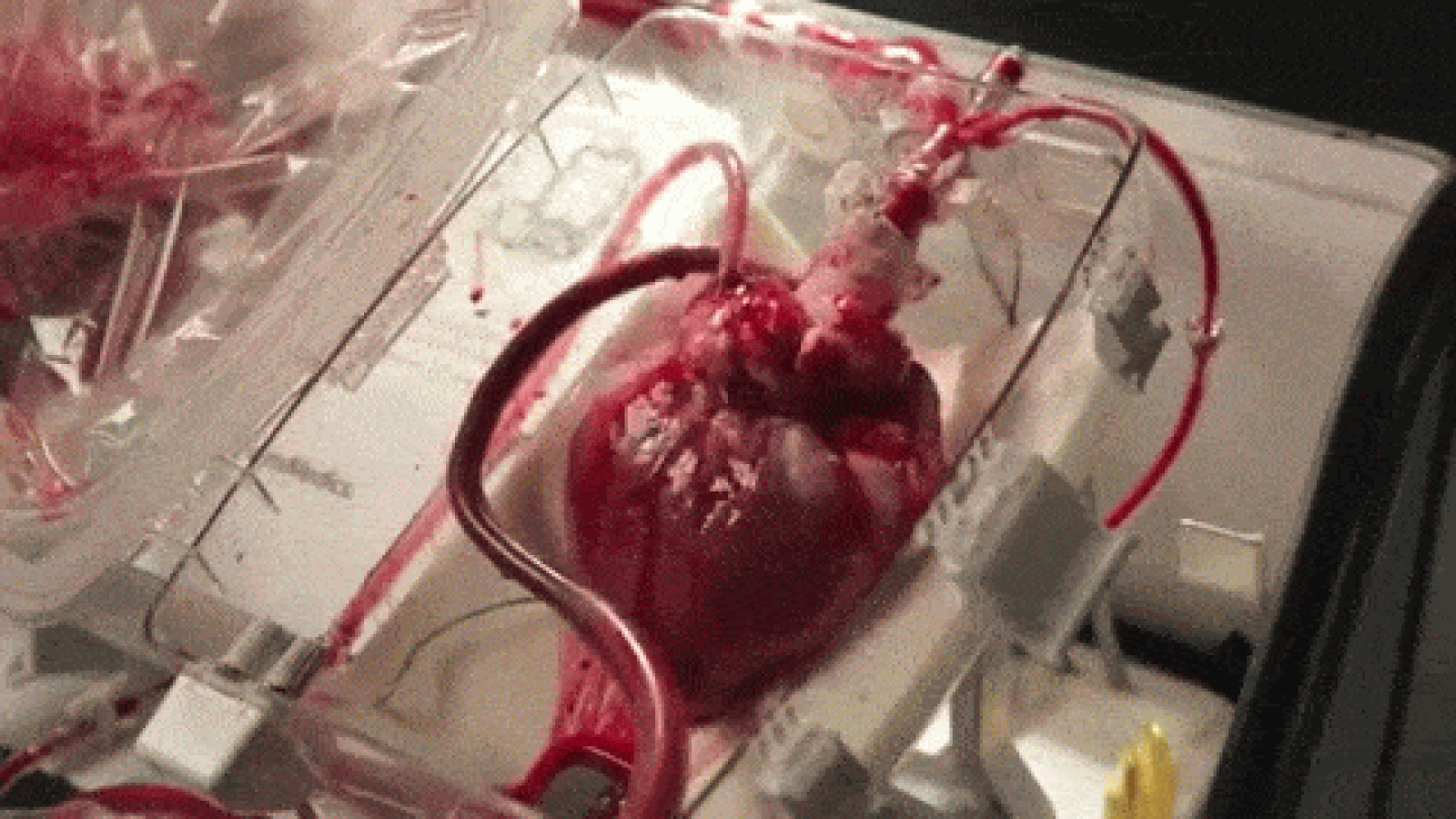
A device that reanimates organs taken from dead patients has shown promise in heart transplant surgeries, though it’s raising some ethical concerns, as well. As MIT Technology Review reports, the so-called “heart in a box” uses tubing and oxygen to pump blood and electrolytes into hearts from recently deceased patients, allowing the organs to continue functioning within a chamber. The system, developed by Massachusetts-based Transmedics, has been successfully deployed in at least 15 heart transplants in the UK and Australia, and is awaiting regulatory approval in the US.
Until now, hearts used for transplants have usually been extracted from brain-dead patients; those from dead patients have been considered too damaged. Once removed, the hearts are also stored and transported in cold temperatures to avoid rapid deterioration, though scientists have begun using devices like the heart in a box to keep the organs warm and functioning. That, doctors say, could increase the pool of donated hearts by between 15 and 30 percent.
Some say the $250,000 device is still too expensive to be deployed widely, and that it needs greater automation. For medical ethicists, the question is how long surgeons should wait before removing a heart that has stopped. “How can you say it’s irreversible, when the circulatory function is restored in a different body?” Robert Truog, an ethicist at Harvard University, tells MIT Technology Review. “We tend to overlook that because we want to transplant these organs.” Truog says he believes those patients can be considered dead, though it’s ultimately a decision for family members to make. “They are dying and it’s permissible to use their organs. The question is whether they are being harmed, and I would say they are not.”