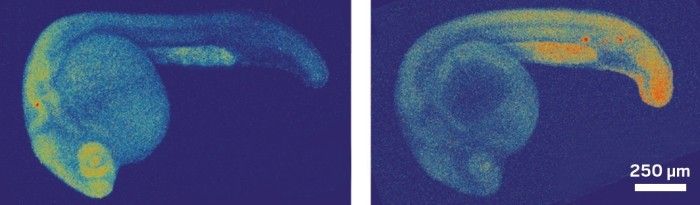
The ability to track molecular events inside the cells of living organisms offers a powerful window into fundamental biological processes, but methods for visualizing RNA in vivo without interfering with cell processes have been elusive. Now, researchers have developed a light-induced chemical reaction that accomplishes this feat in live zebrafish embryos (ACS Cent. Sci. 2016, DOI: 10.1021/acscentsci.6b00054). It is the first technique for detecting specific strings of nucleic acids in live vertebrates that doesn’t require genetically modifying the organism. What’s more, it’s sensitive enough to visualize the expression of microRNAs, small noncoding RNAs that act as puppetmasters of gene expression.
To do the reaction, chemical biologist Nicolas Winssinger, biochemist Marcos Gonzalez-Gaitan, and their colleagues at the University of Geneva designed two nucleic acid probes that each complement and bind to adjacent halves of a target microRNA sequence. The researchers conjugated one probe to a ruthenium complex that absorbs visible light and the other to a fluorogenic rhodamine that lights up when its azide bonds are cleaved. When the probes attach to the target sequence, the two reagents come close enough to react. Shining a light on the sample activates the ruthenium which then reduces the azide in the rhodamine conjugate, releasing its fluorescence. The dependence on external light allows researchers to control when the reporting reaction happens, Winssinger explains.
The team first developed the system three years ago (Chem. 2013, DOI: 10.1002/chem.201300060) for use in cultured cells; here, they adapted it for use in just-fertilized zebrafish embryos. “That’s really not trivial,” says Winssinger. The probes had to be nontoxic, stable for a day or more, and powerful enough to work even after being diluted through cell division.