https://youtube.com/watch?v=vCQm_2JgLbk
DeepMind CEO and co-founder. “We believe this work represents the most significant contribution AI has made to advancing the state of scientific knowledge to date. And I think it’s a great illustration and example of the kind of benefits AI can bring to society. We’re just so excited to see what the community is going to do with this.” https://www.futuretimeline.net/images/socialmedia/
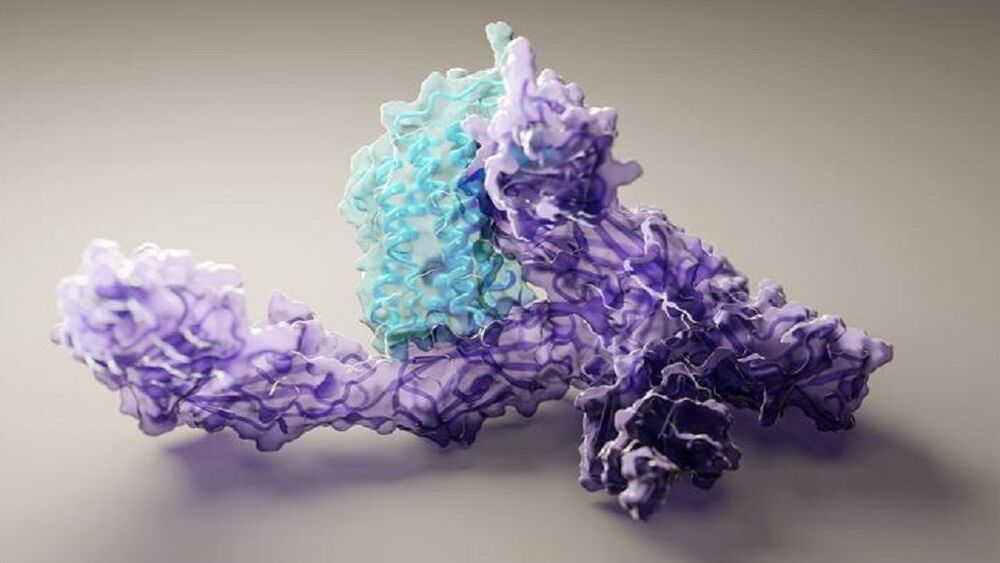
AlphaFold is an artificial intelligence (AI) program that uses deep learning to predict the 3D structure of proteins. Developed by DeepMind, a London-based subsidiary of Google, it made headlines in November 2020 when competing in the Critical Assessment of Structure Prediction (CASP). This worldwide challenge is held every two years by the scientific community and is the most well-known protein modelling benchmark. Participants must “blindly” predict the 3D structures of different proteins, and their computational methods are subsequently compared with real-world laboratory results.
The CASP challenge has been held since 1994 and uses a metric known as the Global Distance Test (GDT), ranging from 0 to 100. Winners in previous years had tended to hover around the 30 to 40 mark, with a score of 90 considered to be equivalent to an experimentally determined result. In 2018, however, the team at DeepMind achieved a median of 58.9 for the GDT and an overall score of 68.5 across all targets, by far the highest of any algorithm.
Then in 2020, version 2.0 of their AlphaFold program competed in the CASP, winning once again – this time with even greater accuracy. The AlphaFold 2.0 achieved a median of 92.4 across all targets, with its average margin of error comparable to the width of an atom (0.16 nanometres). Andrei Lupas, biologist at the Max Planck Institute in Germany who assessed the performances of each team in CASP, said of AlphaFold: “This will change medicine. It will change research. It will change bioengineering. It will change everything.”