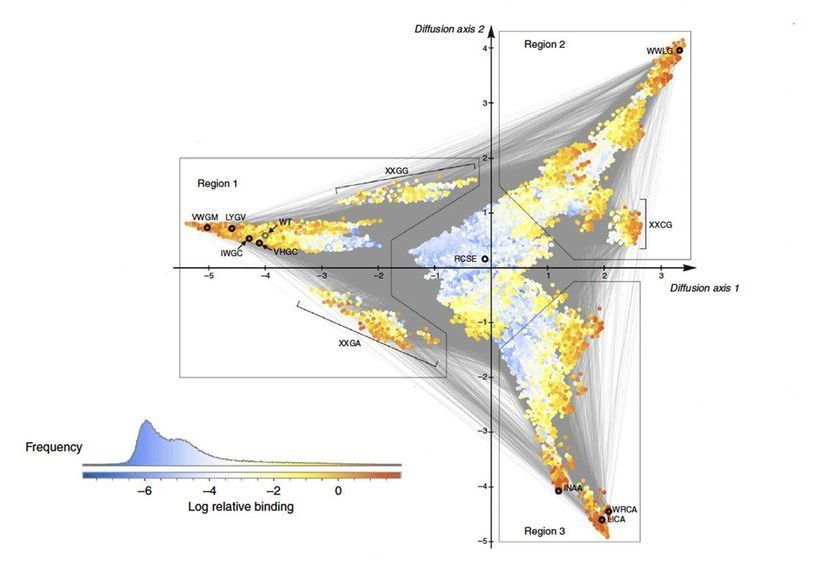
Quantitative biologists David McCandlish and Juannan Zhou at Cold Spring Harbor Laboratory have developed an algorithm with predictive power, giving scientists the ability to see how specific genetic mutations can combine to make critical proteins change over the course of a species’ evolution.
Described in Nature Communications, the algorithm called “minimum epistasis interpolation” results in a visualization of how a protein could evolve to either become highly effective or not effective at all. They compared the functionality of thousands of versions of the protein, finding patterns in how mutations cause the protein to evolve from one functional form to another.
“Epistasis” describes any interaction between genetic mutations in which the effect of one gene is dependent upon the presence of another. In many cases, scientists assume that when reality does not align with their predictive models, these interactions between genes are at play. With this in mind, McCandlish created this new algorithm with the assumption that every mutation matters. The term “Interpolation” describes the act of predicting the evolutionary path of mutations a species might undergo to achieve optimal protein function.